Abstract
Cytogenetically normal acute myeloid leukaemia (CN-AML) accounts for approximately 25%-30% of paediatric AML cases and carries a high risk of relapse. Minimal residual disease (MRD) is an essential factor in predicting relapse in acute leukaemia but is difficult to track for many CN-AML patients, due to the lack of a distinct and stable molecular marker. Consequently, new biomarkers are urgently required for MRD monitoring of the disease. Splicing variants, products of another hallmark of human cancers, aberrant splicing, have been shown informative in predicting responses to cancer treatment. Therefore, we characterized splicing events according to different cytogenetic features by targeted RNA-seq and interrogated the use of splicing variants in MRD monitoring of CN-AML.
A total of 29 AML samples, collected from 18 de novo paediatric AML patients (median age 5.66 years, range 0.67 - 16.38 years) were analysed for this study. Among the 29 samples, 52% harboured a chromosome translocation, 21% were cytogenetically normal, and 28% showed a complex karyotype (defined as having 3 or more cytogenetic features). 100ng of total RNA, extracted from peripheral blood or bone marrow were subjected to library preparation using the Archer™ FusionPlex™ Heme and Myeloid panels, then sequenced using Illumina MiSeq® or NextSeq®. Novel splicing events and genetic mutations were identified by the Archer Dx analysis software in conjunction with normalisations against the library size and probe numbers. Splicing variants were validated using splicing junction-specific probe assays.
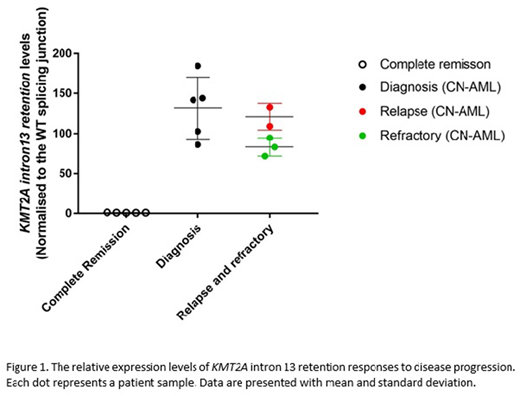
Our results revealed 3249 novel splicing events in 29 AML samples. These events were classified into 4 major types (65% intron retention, 10% exon skipping, 8% exon out of order and 8% intra-exon gap), and 9 minor events that were combinations of the major types (9%). The number of splicing events per sample was not associated with the disease status or the presence of the mutations in the spliceosome encoding gene, SF3B1 or U2AF1. Instead, splicing variants were associated with cytogenetic features. Of note, an intron 13 retention of the KMT2A (MLL) gene was identified in all CN-AML samples, and was consistently expressed approximately 100 times higher in the CN-AML compared to other AML cases or remission samples. To assess whether KMT2A intron 13 retention could be a potential molecular MRD marker to monitor CN-AML, we measured its expression in samples from 5 independent CN-AML patients who had available samples for 3 time-points of disease progression. Our results demonstrated, with 95% detection power, that KMT2A intron 13 retention was differentially expressed at different time points (Figure 1). Moreover, the expression level of this splicing variant correlated with disease progression in every patient examined. In conclusion, these data suggest that intron 13 retention of KMT2A may be a novel molecular marker for MRD monitoring in CN-AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal